Phylogenetic Tree of Catalase
Did you know that you share almost 75% of your most important genes with fruit flies? We are all more similar than it seems. This activity will help explore just how similar different animals are.
Our goal today is to take a collection of sequences and proteins, and compute a phylogentic tree based on the most likely sequence of mutations that would produce these observed species. When this is complete, we will also infer what species are more closely related.
Catalase
Next consider the enzyme catalase. From Wikipedia:
Catalase is a common enzyme found in nearly all living organisms exposed to oxygen (such as vegetables, fruit or animals). It catalyzes the decomposition of hydrogen peroxide to water and oxygen.
That is, Catalase is a protein that catalyzes this reaction: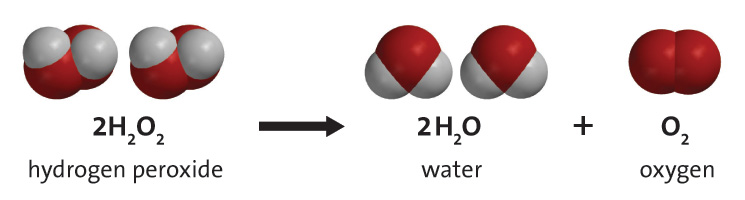
Like all proteins, Catalase is encoded by a messenger RNA. Here you will find the mRNA sequences for Catalase: Catalase mRNA sequences
for the following animals:dog mouse human horse chimp platypus tazmanian devil gorilla elephant cat ferret panda megabat bushbaby
Before we make a phylogenetic tree for this mRNA, make some guesses about which animals you think will be most closely related. If you don’t know what an animal looks like, search for it in Google Images. For each of the following animals, which of the two other animals will be more similar?:
Mouse: is it closer to Elephant or Ferret?
Platypus: is it closer to Dog or Tasmanian Devil?
Horse: is it closer to Elephant or Panda?
Bushbaby: is it closer to Chimp or Mouse?
Panda: is it closer to Ferret or Cat?
Phylogenetic Tree from mRNA sequences
Let's try to create a phylogenetic tree! Open the mRNA sequences here: Catalase mRNA sequences
Copy and Paste the three animals you want to compare. For example, for the first three we have mouse, elephant, and ferret. The resulting sequenes should look like this:
>mouse catalase GAAGTCACCACTCCAGCGGGCCTGGCCAACAAGATTGCCTTCTCCGGGTGGAGACCGCTGCGTCCGTCCCTGCTGTCTCACGTTCCGCAGCTCTGCAGCTCCGCAATCCTACACCATGTCGGACAGTCGGGACCCAGCCAGCGACCAGATGAAGCAGTGGAAGGAGCAGCGGGCCTCGCAGAGACCTGATGTCCTGACCACCGGAGGCGGGAACCCAATAGGAGATAAACTTAATATCATGACCGCGGGGTCCCGAGGGCCCCTCCTCGTTCAGGATGTGGTTTTCACTGACGAGATGGCACACTTTGACAGAGAGCGGATTCCTGAGAGAGTGGTACACGCAAAAGGAGCAGGTGCTTTTGGATACTTTGAGGTCACCCACGATATCACCAGATACTCCAAGGCAAAGGTGTTTGAGCATATTGGAAAGAGGACCCCTATTGCCGTTCGATTCTCCACAGTCACTGGAGAGTCAGGCTCAGCTGACACAGTTCGTGACCCTCGGGGGTTTGCAGTGAAATTTTACACTGAAGATGGTAACTGGGATCTTGTGGGAAACAACACCCCTATTTTCTTCATCAGGGATGCCATATTGTTTCCATCCTTTATCCATAGCCAGAAGAGAAACCCACAGACTCACCTGAAGGATCCTGACATGGTCTGGGACTTCTGGAGTCTTCGTCCCGAGTCTCTCCATCAGGTTTCTTTCTTGTTCAGTGACCGAGGGATTCCCGATGGTCACCGGCACATGAATGGCTATGGATCACACACCTTCAAGTTGGTTAATGCAGATGGAGAGGCAGTCTATTGCAAGTTCCATTACAAGACCGACCAGGGCATCAAAAACTTGCCTGTTGGAGAGGCAGGAAGGCTTGCTCAGGAAGATCCGGATTATGGCCTCCGAGATCTTTTCAATGCCATCGCCAATGGCAATTACCCGTCCTGGACGTTTTACATCCAGGTCATGACTTTTAAGGAGGCAGAAACTTTCCCATTTAATCCATTTGATCTGACCAAGGTTTGGCCTCACAAGGACTACCCTCTTATACCAGTTGGCAAACTGGTTTTAAACAAAAATCCAGTTAATTACTTTGCTGAAGTTGAACAGATGGCTTTTGACCCAAGCAATATGCCCCCTGGCATCGAGCCCAGCCCTGACAAAATGCTTCAGGGCCGCCTTTTTGCCTACCCGGACACTCACCGCCACCGCCTGGGACCCAACTATCTGCAGATACCTGTGAACTGTCCCTACCGCGCTCGAGTGGCCAACTACCAGCGTGATGGCCCCATGTGCATGCATGACAACCAGGGTGGTGCCCCCAACTATTACCCCAACAGCTTCAGCGCACCAGAGCAGCAGCGCTCAGCCCTGGAGCACAGCGTCCAGTGCGCTGTAGATGTGAAACGCTTCAACAGTGCTAATGAAGACAATGTCACTCAGGTGCGGACATTCTACACAAAGGTGTTGAACGAGGAGGAGAGGAAACGCCTGTGTGAGAACATTGCCGGCCACCTGAAGGACGCTCAGCTTTTCATTCAGAAGAAAGCGGTCAAGAATTTCACTGACGTCCACCCTGACTATGGGGCCCGCATCCAGGCTCTTCTGGACAAGTACAACGCTGAGAAGCCTAAGAACGCAATTCACACCTACACGCAGGCCGGCTCTCACATGGCTGCGAAGGGAAAAGCTAACCTGTAACTCCGGTGCTCAGCCTCCGCTGAGGAGACCTCTCGTGAAGCCGAGCCTGAGGATCACCTGTAATCAACGCTGGATGGATTCTCCCACTCCGGAGCGCAGACTCACGCTGATGACTTTAAAACGATAATCCGGGCTTCTAGAGTGAATGATAACCATGCTTTTGATGCCGTTTCCTGAAGGGAAATGAAAGGTTAGGGCTTAGCAATCATTTAACAGAAACATGGATCTAATAGGACTTCTGTTTGGATTATTCATTTAAATGACTACATTTAAAATGATTACAAGAAAGGTGTTCTAGCCAGAAACATGACTTGATTAGACAAGATAAAAATCTTGGCGAGAATAGTGTATTCTCCTATTACCTCATGGTCTGGTATATATACAATACAACACACATACCACACACACACACACATGCAATACACACACTACACACACATACACACACTCACACACACTCATACACACACATGAAGAGATGATAAAGATGGCCCACTCAGAATTTTTTTTTTTATTTTTCTAAGGTCCTTATAAGCAAAACCATACTTGCATCATGTCTTCCAAAAGTAACTTTAGCACTGTTGAAACTTAATGTTTATTCCTGTGCTGTGCGGTGCTGTGCTGTGCTGTGCTGTGCAGCTAATCAGATTCTTGTTTTTTCCCACTTGGATTATGTTGATGTTAATACGCAGTGATTTCACATAGGATGATTTGTACTTGCTTACATTTTTACAATAAAATGATCTACATGGAAGGACCGTGTTTGGTTGCTTTCAGCTCTGTATAATGTGGAATGTGAAGTAGAGATTACCAGCTCTCTCTGCAGTAACAATAAAAGCGCCAGCGGCCAGA >elephant catalase CTGTTTAGGGTGAAGACGTCTGACCTGAGGCAGCCTGCAAGGTTCTGCAACCCAACTCGGACACCATGGCGGACAGCCGGGACCCAGCCAGCGACCAGATGAAGCGCTGGAAGGAGGAGCGTTCCACGCAGAGACCTAGTGTCCTGACCACTGGAGCCGGCAACCCTGTAGGAGACAAACTTAACTCTATGACAGTAGGGCCCCGGGGACCCCTTCTGGTTCAGGATGTGGTTTTCACTGATGAAATGGCTCACTTTGACCGAGAGAGAATTCCTGAGAGAGTCGTGCATGCTAAAGGAGCAGGGGCCTTTGGCTACTTTGAGGTCACACATGACATTACCAGATACTCCAAGGCAAAGGTGTTTGAGCATATTGGAAAGAGGACTCCCATTGCAGTTCGATTCTCCACTGTTGCTGGAGAATTGGGCTCAGCTGACACAGTTCGTGACCCTCGGGGGTTTGCAGTGAAATTTTACACAGAAGATGGTAACTGGGATCTCGTTGGAAATAACACTCCCATTTTCTTCATCAGGGATGCCTTATTGTTCCCCTCCTTTATCCACAGCCAAAAGAGAAACCCTCAAACGCACCTGAAGGATCCCGACATGGTGTGGGACTTCTGGAGCCTGCGCCCCGAGTCTCTGCATCAGGTTTCTTTCCTGTTCAGTGATAGAGGGATTCCAGACGGACATCGGCACATGAATGGATATGGATCACATACTTTCAAACTGGTCAATGCAGACGGAGAGGCAGTTTATTGCAAATTCCATTATAAGACTGACCAAGGCATCAGAAACCTTTCTGTGGAAGATGCAGCAAGACTTTCCCAGGAAGATCCTGACTATGGCATCCGAGATCTTTATAATGCCATTGCCACAGGCAACTACCCCTCCTGGACCTTTTACATCCAGATCATGACATTCAGTCAGGCAGAAAATTTTCCATTTAATCCATTTGATCTCACCAAGATTTGGCCTCACAAGGACTTTCCTCTTATTCCAGTTGGTAAACTGGTCTTAAACCGGAACCCTGTCAATTACTTTGCTGAGGTTGAACAAATAGCCTTTGACCCAAGCAACATGCCACCCGGCATTGAGCCCAGCCCTGACAAAATGCTTCAGGGCCGTCTTTTTGCCTATCCTGACACTCACCGCCACCGCCTGGGCCCCAACTACCTTCAGATACCTGTGAACTGTCCCTACCGTGCTCAGGTGGCCAACTACCAGCGTGATGGCCCCATGTGTATGCTGCACAACCAAGGTGGTGCCCCAAATTACTACCCCAACAGCTTCAGCGCCCCGGAACAACAGCGCTCTGCCCTGGAGCACGGCACCCGCTGTTCTGGGGATGTGCAGCGCTTCAACAGTGCTGACGAGGACAACGTCACTCAGGTGCGAACATTCTATAAAAACGTGCTGAATGATGAACAGAGGCAACGCCTGTGTGAGAACATTGCAGGCCATCTGAAAGACGCACAGCTTTTTATCCAGAAGAAAGCGGTCAAGAACTTCAGTGATGTCCACCCTGACTACGGGGCCTGCATCCAGGCGCTTCTGGACAAATACAACGCTGAGAAACCTAAGAACGCGATCCACTCCTTTGTGCAGCATGGGTCTCACTTGGCCGCGAGGGAAAAGGCTAACCTGTGAGGTCTGGGGCCTGCGTCCTTGGCTGCGAAGCTCTCAGTCCATGAAGCAAAGCATGACGTCCACACGTGCCATGTTCTTATCGCTGGATAGGAGGTTCTCCTGTGCTGGGTGTGCAAATGCAAGCTAATGCCTTTAACATGGTAATTCAGGTTTCTGTAGCAAATAACCTAATGATAACTTTTCATAGTTTTAATAACTTTTAATTATTTCCCCTAGGGAAATTGGGGGTAAATGGAGGTTAGGGCTTAATAAGCGTTTAAAAGAAACATATACTGGCTTTTGACAGTAAGTTGGATTATTCATTTAAAATGACTAGAAGGAAAGTTTCTAGCAGAAATATGATTTTATTTGGTAAGAAAAAAAATCTTGGTTAAAATTGGTGTGTTTACAAATTACCTCATGGCCTATTAAATAAAATCATGGCTATAATGATACAAGAAGAAAAAAAGATGATCCACCTGGACATTTTTATTTTTATAAATTCTTTGTAGGAAAAACACCTTTAATACATCAGTGCCTTCCAAAAAGAACTTGAGCCACCATCATGGCTTAATGTGTATTCCAGCTTGGAATTGATCAACTTTTTGTTTTTCTCTTGGAATCATATTGATTACATACAGCACTGATTTTGCAACAGGCTGATAGGTAATTGCTTACATTGTTACAATAAAGTAAT >ferret catalase GGCGGAGTGACAGGCGGAGGCAGAAGTCGCCTACTTTATGTCGCGCGGCGGTAGTCGGCGTCGTCCGCTGAGGGTGGAGACGTGAGAACCGAGGCCACCTGCAACGTTCTGCAAAGCCAGTCACACACGATGGCGGACAGCCGGGATCCAGCCAGCGACCAGATGAAGTTCTGGAAGGAGCAGCGGGCCGCGCAGAAACCCGATGTCCTGACCACTGGTGCCGGTAATCCAGTCGGAGACAAACTCAATGTTATGACATCAGGGCCCCGGGGTCCCCTTCTCGTTCAGGATGTGGTATTCACCGATGAAATGGCTCACTTTGACCGGGAAAGAATCCCTGAGAGAGTCGTGCACGCCAAAGGAGCAGGGGCTTTTGGCTACTTCGAGGTCACTCATGACATTACCAGATACTCCAAAGCGAAGGTGTTTGAGCATATTGGAAAGAGGACTCCCATTGCTGTTCGATTCTCCACTGTCGCTGGAGAGTCAGGCTCAGCGGACACAGTTCGGGACCCTCGTGGGTTTGCTGTGAAATTTTACACAGAGGATGGTAATTGGGATCTTGTTGGAAATAACACCCCCATTTTCTTCATCAGGGATGCCATATTGTTTCCATCCTTTATCCATAGTCAAAAGAGAAACCCTCAAACACACCTGAAGGATCCCGACATGGTCTGGGACTTCTGGAGCCTGCGCCCCGAGTCTCTGCATCAGGTTTCCTTCCTGTTCAGTGATCGAGGGATTCCAGATGGACACAGGCACATGAACGGATACGGATCACATACTTTTAAGCTGATCAATGCGAAGGGAGAGGCAGTTTATTGCAAATTCCATTATAAGACTGACCAGGGCATCAAAAACCTTTCTGTGGAAGACGCTGCAAGACTTTCTCAGGAAGATCCTGACTACAGCCTGCGGGATCTTTTCAATGCCATTGCCACGGGCAACTACCCCTCCTGGACATTTTACATCCAGGTCATGACTTTTAATCAGGCAGAAACCTTTCCATTTAATCCATTTGATCTTACCAAGATTTGGCCTCACCAGGACTATCCTCTTATCCCAGTTGGTAAACTGGTCTTAAACCGGAATCCAGTTAATTACTTTGCTGAGGTTGAACAGTTGGCATTTGACCCAAGCAACATGCCACCTGGCATTGAGCCCAGTCCTGACAAAATGCTTCAGGGCCGCCTTTTTGCCTATCCTGATACTCACCGCCACCGCCTGGGACCCAACTATCTTCAGATACCTGTGAACTGTCCTTTCCGTGCTCGAGTGGCCAACTACCAGCGTGACGGCCCCATGTGCATGCTGGACAATCAGGGTGGTGCTCCAAATTACTACCCCAATAGTTTTAGTGCTCCAGAACAGCAGCCTTCTGCCCTGGAACATAGCAGCCAGTGCTCTCCAGACGTGCAGCGCTTCAACAGTGCCAACGAAGATAATGTCACTCAGGTGCGGACGTTCTACACGAAGGTGCTGAATGAGGAGGAGAGGAAACGCCTGTGCAAGAACATTGCGGGCCACCTGAAGGATGCACAGCTTTTCATCCAGAAGAAAGCGGTCAAGAACTTCAGTGATGTCCATCCTGACTATGGAGCACGCATCCAGGCTCTTTTGGACGAATACAATGCTCAGAAACCCAAGAATGCGATTCACACCTTTATGCAGCATGGGTCCCACCTGGCGGCAAGGGAGAAAGCCAACCTGTGAGTCGGGGCCCCGGGCCTGCCCCAAGTTGCTCTCCATCTGAGAAGCAAACCATGGTGTTCACACACCTACCCACTCTTTGCCAGATAGAAGATTCTCCTGGGCTAGTTGCACAATCGCAAGCCATGTCTTTCAAATAATAATCCAGGTTTCTATCGCAAATAACGTAGCAGTGGCGTTTAGCGCTATTTCCCTGGGGGGGAATAAGGGTAGGGCTTAATAGTGGTAAAAAAGAAAACGTACTTGCTTTTGACAGTTGATTGGATTATTCACTTAACATGACTAGAATGACAGTTTCTGGCAGAAATATGATTTTATTTGATGAGAAGAAAATCTTGGTGAAATTAGTATGTTTACATATCATCTCATGGCCTTATTGTATAAAAATATGGCTGTAATTGTATAAGAAGAAAAGATCACCTACTCAGTAATTTTCATTTCTCTCAGTTCTGTATAGGAAAAACACATTTAATGCATTGATGTCTTTTGAAAATAATTTCACTGACATAATAGCTTAATGCTTACTCCTACCTGGAACTGAACTTGGAATTACATCTATGCTCATATAGCATTGATTTTGCAACAGACTGATTTGTAATTGCTACATTTTTACAATAAAATAATCTGCACATAAGAA
Create a multiple sequence alignment with Clustal Omega. Click the "Catalase mRNA sequences" link below into a new tab, and paste the sequences here: Clustal Omega. Make sure you select RNA from the menu. and then click "Submit" at the bottom of the page.
Now View the phylogenetic tree. Once the job is done, you can view the resulting sequence alignment from Clustal Omega. This alignment is computed to arrange the amino acids to make the similarty between sequences as good as possible. Next select the Phylogenetic Tree tab from above. Then scroll down to the tree. Which one of your predictions was correct? Which was incorrect?
Let's say you don't like the style of tree drawn by this program. An important idea of computational biology software, is that research is done in steps, in which case the software is said to be modular. For example, you can create your own tree image using the data from this page. From within the same Phylogenetic Tree tab, above the tree is a representation of the tree in Newick Tree format. You can create a new, different tree using T.Rex, by copying and pasting it here: T.Rex Tree Viewer. Select "Reset" to clear the input, paste the Newick Tree Format into the box, and click "View Tree"
mRNA vs Protein:
Now let's try this with protein sequences. You can find the sequences of the Catalase (protein sequences) here Catalase protein sequences. It contains catalase protein sequences for the same animals as before.
How does the result differ when you use the protein sequences intead of the mRNA sequences?
Paste into Clustal Omega and build another tree. This time, choose Protein as the sequence type.
.Ribosomal RNA
Now let's examine Ribosomal RNA sequences. Ribosomal RNAs (rRNAs) are transcribed, but are not translated into a protein. These RNAs have many important biological roles. In fact, rRNAs are involved in translation, even though they are not translated!
From wikipedia:In molecular biology, ribosomal ribonucleic acid (rRNA) is the RNA component of the ribosome, and is essential for protein synthesis in all living organisms. It constitutes the predominant material within the ribosome, which is approximately 60% rRNA and 40% protein by weight....The ribosomal RNAs form two subunits, the large subunit (LSU) and small subunit (SSU).
You can download a file of 18S (part of the small subunit) ribosomal RNAs here: 18S rRNAs It contains the 18S RNA sequences for the following organisms:
A protozoan parasite called Sarcocystis cruzi Millipede Earthworm Horseshoe crab Starfish A Flea. A species found in Korea related to the snowflea. See some images here: Hypogastrura dolsana Rabbit A type of free-living (not parasitic) microscopic "flatworm" called a Triclad Corn Plant Cucumber Poplar Tree Fruit Fly Mouse
Before we make a phylogenetic tree for this RNA, how do you think it will turn out? Make some guesses on which organism is more closely related to the first:
Flea: do you think it's closer to Fruit Fly or to Horseshoe crab
Earthworm: do you think it's closer to the Starfish or to the Flatworm (the "Triclad").
Cucumber: do you think it's closer to the Corn Plant or to the Poplar Tree.
Create a multiple sequence alignment using Clustal Omega. Open the 18S rRNAs into a new tab, and pasted the sequences and deflines from the fasta file into this web program here: Clustal Omega. Make sure you select RNA from the menu.
Now View the phylogenetic tree. Once the job is done, you can view the resulting sequence alignment from Clustal Omega. This alignment is computed to arrange the nucleotides to make the similarty between sequences as good as possible. Next select the Phylogenetic Tree tab from above. Then scroll down until you see a phylogentic tree like shown before on the slides.
You can optionally create your own tree image by copying and pasting the Newick Tree into T-rex as you did before here: T.Rex Tree Viewer.